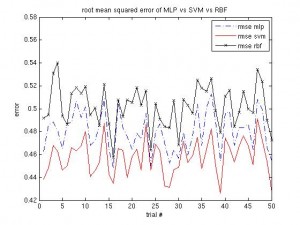
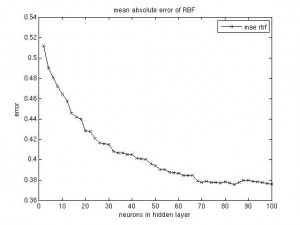
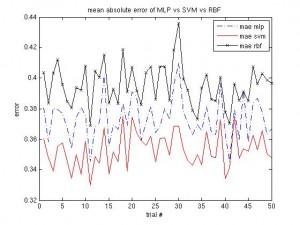
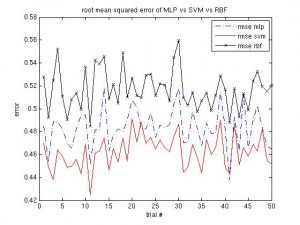
It’s been a long time since the last post. Same procedure as every year: once the winter term starts, the teaching stress starts again. About two weeks into the term I caught an RSI syndrome (http://en.wikipedia.org/wiki/Repetitive_strain_injury) that prevented me from any typing, mousing and writing activities. It certainly feels better now, but I was given advice by three doctors to not overuse my hands any more, otherwise it would get worse than last time and surgery would be needed. I’m more or less confined to reading literature and taking some notes, as the pain usually returns after a short while of typing.
Nevertheless, I’m in Cambridge, England, at the moment. Yesterday I took part in the FAIRS’08 workshop, which was held at the same location as last year. There were about 20-25 PhD AI research students and seven to ten experienced (seasoned) experts for giving guidance along the PhD path. The best presentation was certainly given by Max Bramer, followed closely by Frans Coenen. Further scholars were Adrian Hopgood, Simon Thompson, Miltos Petridis, among others. Many thanks to Alice Kerly who perfectly organised the full forum day.
I gave my presentation on what I’m planning to do in my PhD, too. It raised some interdisciplinarity issues, i.e. I will have to take care that I’ll be doing a computer science PhD, not an agriculture PhD. At least my motivation was quite clear — I’m interested in my topic and it’s fun working on it. My slides are here:slides-2008fairs